基因编辑技术可以对生物基因组进行精准的修饰,开发针对编辑后基因型的简单易用的检测方法对于该技术在作物(尤其在多倍体作物)中的大规模应用至关重要。最近,中科院遗传所高彩霞团队开发了一个名为PCR/RNP的检测方法,显示出了极大的应用前景。
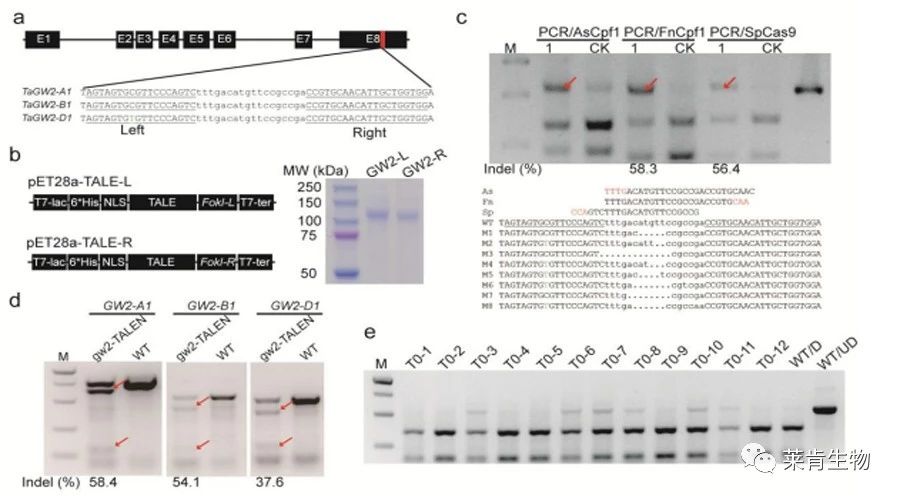
研究人员应用纯化的CRISPR核糖核蛋白复合体消化靶标区域的PCR产物,如果靶标区域成功编辑,该区域的原有的gRNA发生变化,核糖核蛋白复合体将不能切割PCR片段。研究者们运用该方法检测六倍体小麦和二倍体水稻中的编辑突变体,结果显示,这种检测方法相比Sanger测序更灵敏,比PCR/RE方法更易用(因为不需要酶切位点),比二代测序更便宜。由于靶标位点周围布满大量的SNP,该方法尤其适合对编辑后的小麦基因型进行检测。此外,该方法也可以对纯化的TALEN蛋白诱导的DNA-free编辑以及基因组单碱基替换进行检测。
PCR/RNP检测方法简单易用、成本低、限制少、适用性广,将成为检测编辑作物基因型的有力工具。
Plant Biotechnol J. 2018 May 3.
Genotyping genome-edited mutations in plants using CRISPR ribonucleoprotein complexes.
Author
Zhen Liang, Kunling Chen, Yan Yan, Yi Zhang, Caixia Gao*
*: State Key Laboratory of Plant Cell and Chromosome Engineering, Center for Genome Editing, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, China.
Abstract
Despite the great achievements in genome editing, accurately detecting mutations induced by sequence-specific nucleases is still a challenge in plants, especially in polyploidy plants. An efficient detection method is particularly vital when the mutation frequency is low or when a large population needs to be screened. Here, we applied purified CRISPR ribonucleoprotein complexes to cleave PCR products for genome-edited mutation detection in hexaploid wheat and diploid rice. We show that this mutation detection method is more sensitive than Sanger sequencing and more applicable than PCR/RE method without the requirement for restriction enzyme site. We also demonstrate that this detection method is especially useful for genome editing in wheat, because target sites are often surrounded by single nucleotide polymorphisms. By using this screening method, we were also able to detect foreign DNA-free tagw2 mutations induced by purified TALEN protein. Finally, we show that partial base editing mutations can also be detected using high fidelity SpCas9 variants or FnCpf1. The PCR/RNP method is low-cost and widely applicable for rapid detection of genome-edited mutation in plants.